Publications
Knowledge of the variation and distribution of substrates at large spatial extents in aquatic systems, particularly rivers, is severely lacking, impeding species conservation and ecosystem restoration efforts. Recreation-grade side-scan sonar (SSS) instruments have demonstrated their unparalleled value as a low-cost scientific instrument capable of efficient and rapid imaging of the benthic environment. However, existing methods for generating georeferenced data sets from these instruments, especially substrate maps, remain a barrier of adoption for scientific inquiry due to the high degree of human intervention and required expertise. To address this shortcoming, we introduced PING-Mapper, an open-source and freely available Python-based software for automatically generating geospatial benthic data sets from popular Humminbird® instruments reproducibly. The previously released Version 1.0 of the software provided automated workflows for exporting georeferenced sonar imagery. This study extends functionality with version 2.0 by incorporating semantic segmentation with deep neural networks to reproducibly map substrates at large spatial extents. We present a novel approach for generating label-ready sonar data sets, creating label-image training sets, and model training with transfer learning; all with open-source tools. The six-class substrate model achieves an overall accuracy of 78% and the best performing class achieves 91%. Grouping substrates into three classes further improves the overall accuracy (87%) and best performing class accuracy (94%). Additional workflows enable masking sonar shadows, calculating independent bed picks and correcting attenuation effects in the imagery to improve interpretability. This software provides an improved mechanism for automatically mapping substrate distribution from recreation-grade SSS systems, thereby lowering the barrier for inclusion in wider aquatic research.

The characterization of benthic habitats is essential for aquatic ecosystem science and management, but is frequently limited by waterbody visibility and depth. Recreation‐grade side‐scan sonar systems are increasingly used to aid scientific inquiries in aquatic environments due to their relative low‐cost, ease of operation, low‐weight, and ease of mounting on a variety of vessels. However, existing procedures and software for post‐processing these data are either limited, closed source, or fail on data from new sonar models, limiting the development of reproducible workflows. Here, we present PING‐Mapper, an open‐source and freely available side‐scan sonar post‐processing toolset for processing and mapping sonar recordings from popular Humminbird instruments. The modular software automatically: (a) decodes sonar recordings from any Humminbird system, (b) exports ping attributes from every sonar channel, (c) uses sonar sensor depth for water column removal, and (d) exports sonogram tiles and georectified mosaics. Sonar channels are processed in parallel for quick decoding and metadata extraction. Major processing wokflows, including georectification and image export, are optimized to scale with computing resources. The software has been extensively tested using data from several rivers of varying character and distribution of depths, but could also be used in estuarine and lacustrine environments. Usage of PING‐Mapper is illustrated in three case studies focused on mapping large woody debris, bathymetric mapping, and visual interpretation and mapping of substrates for selected reaches of the Pearl and Pascagoula river systems in Mississippi.
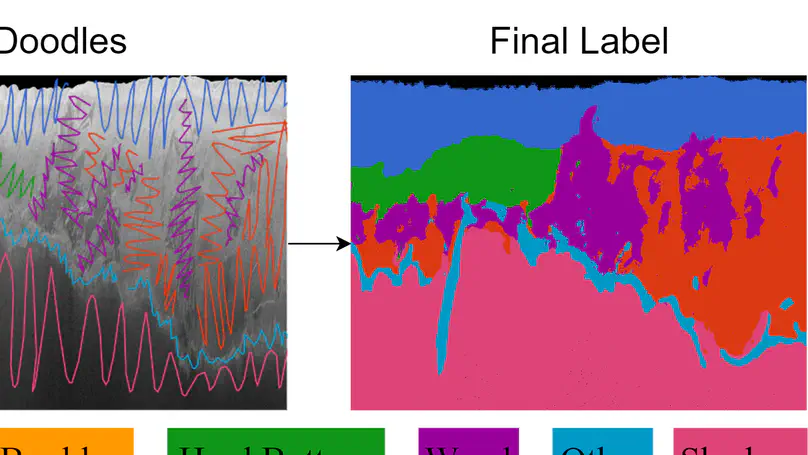
Segmentation, or the classification of pixels (grid cells) in imagery, is ubiquitously applied in the natural sciences. Manual methods are often prohibitively time‐consuming, especially those images consisting of small objects and/or significant spatial heterogeneity of colors or textures. Labeling complicated regions of transition that in Earth surface imagery are represented by collections of mixed‐pixels, ‐textures, and ‐spectral signatures, can be especially error‐prone because it is difficult to reliably unmix, identify and delineate consistently. However, the success of supervised machine learning (ML) approaches is entirely dependent on good label data. We describe a fast, semi‐automated, method for interactive segmentation of N‐dimensional (x, y, N) images into two‐dimensional (x, y) label images. It uses human‐in‐the‐loop ML to achieve consensus between the labeler and a model in an iterative workflow. The technique is reproducible; the sequence of decisions made by human labeler and ML algorithms can be encoded to file, so the entire process can be played back and new outputs generated with alternative decisions and/or algorithms. We illustrate the scientific potential of segmentation of imagery of diverse settings and image types using six case studies from river, estuarine, and open coast environments. These photographic and non‐photographic imagery consist of 1‐ and 3‐bands on regular and irregular grids ranging from centimeters to tens of meters. We demonstrate high levels of agreement in label images generated by several labelers on the same imagery, and make suggestions to achieve consensus and measure uncertainty, ideal for widespread application in training supervised ML for image segmentation.